Previously posted at A Tempting Science on January 12, 2021
Cite as: Benner, S. A. (2021) “The DNA of Alien Life”. Primordial Scoop, 2021, e0124. https://doi.org/10.52400/JKKY2544
Have you ever wondered how Spock can have a human mother and a Vulcan father? How all species in a Tatooine cantina eat the same stuff? Why all aliens look like Hollywood actors with prostheses?
Yes, yes, I know: “Steven, what part of the word ‘fiction’ don’t you understand?”
But for those into “the biology of Star Trek”, the answer is clear. Because all life in the galaxy uses pretty much the same biochemistry. Therefore, they all eat the same stuff, such as amino acids for their proteins. Most importantly, they all have DNA to do their genetics.
The writers of Star Trek tried to rationalize this in “The Chase” (TNG 146th episode). In a Hollywood version of panspermia, the Klingons, Romulans, Cardassians, and humans ran around the galaxy for an hour to find the aliens who seeded spots in the Alpha Quadrant with DNA. The ancestors of Klingons, Romulans, Cardassians, and humans. And that DNA has remained with them ever since in one of the most remarkable examples ever of homoplasy (look it up).

In reality, alien biochemistry is likely quite different from our own.
However, NASA’s search for alien life is biased by the presumption that alien biochemistry will resemble terrestrial biochemistry. Even after we and others, including the National Academy of Sciences, published alternative metabolisms for alternative lifeforms, NASA still designs missions to find life on Mars by looking for … the amino acids that you and I have in our proteins.
What might alien biochemistry actually look like?
Years ago, I tried to answer this question with the tools available to me as a graduate student trained to synthesize new organic molecules. I decided to do something never before tried: synthesize alternative forms of DNA. I wanted to know, by experiment, whether molecules other than standard DNA might do things important in life, like store information, replicate, evolve, and adapt.
For a chemist, DNA is more than an acronym. Rather, it is a molecular structure, a linear strand of building blocks that bind specifically to complementary building blocks from another strand. And chemists know that the specificity of binding in terran DNA follows two rules set out by Watson and Crick 70 years ago.
The first rule requires that the two bound building blocks be complementary in size. Big building blocks must pair was small building blocks.
The second rule requires that the two building blocks be complementary in hydrogen bond interactions. More on this in a later post, but treat today the building blocks as Lego pieces: Holes one building block must match prongs in the other.
Together, these two rules let us draw a simplified picture of how DNA building blocks “fit” together.
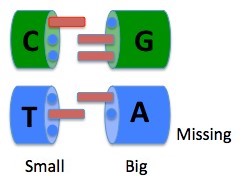
I leave off the chemistry in the picture to make a point. In the DNA C:G pair, C is small, and presents to its partner a prong-hole-hole pattern. Its complement, large G, presents to its partner a hole-prong-prong. Following these two rules of complementarity, DNA strands are held together by C:G pairs.
Natural DNA has two more complimentary building blocks, which work similarly. T is small, and presents to its partner a hole-prong-hole. Its complement, large A, presents to its partner a prong-hole-nothing pattern. Following these two rules of complementarity, DNA strands are held together by T:A pairs.
Even at this level of abstraction, other possibilities are obvious. Why not synthesize a small Lego piece that presents a prong-hole-prong pattern to its large Lego partner that presents a hole-prong-hole pattern? Or a small Lego piece with a hole-hole-prong pattern, and the complementary large Lego partner with a prong-prong-hole pattern?
Here is what these other possibilities look like:
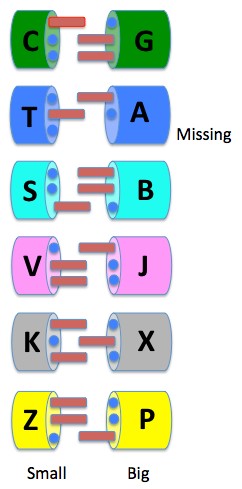
The picture shows an alternative type of DNA not with four building blocks and two independently replicating pairs, but one with as many as 12 building blocks with six independently replicating pairs.
Having conceived of these artificial DNA building blocks, the next step was to actually synthesize them. However, I had a problem. Of all the things in science, it turns out the most important is… money.
As Tom Wolfe wrote, “No bucks, no Buck Rogers”.
There was no way that the National Institutes of Health in the US was going to fund the creation of alien DNA. Fortunately, at just the right moment, the Swiss Federal Institute of Technology offered me a job with funding. I moved to Zürich, and this part of the field of “synthetic biology” was born.
The story has exploded in many ways since then. The alien DNA can store information [1]. It can be copied [2]. It can evolve [3]. And it can adapt [4]. Over $1 billion in medical products [5] have emerged that exploit the properties of alien DNA that we created in Zürich.
So yes, it is possible that Vulcans support their life forms with a kind of genetic molecule that is quite different from what we have in our own bodies. Which means that human-Vulcan hybrids like Spock are… um … fiction.
But before you smugly say: “Steve, yes, we told you”, there is a coda. In 1977, Kimos et al. (not Kodos et al.) reported that a virus infecting a blue green bacterium did not have big A like you and me, with its prong-hole-nothing pattern. No, this virus had decided to “fix” big A to give big A a complete prong-hole-prong pattern. Which now completely matched the hole-prong-hole pattern of small T.
Philippe Marlière went further. Philippe had already shown that terran bacteria could be induced to expand their amino acid repertoires. He then got bacteria to replace T.
The first aliens we are likely to encounter are those that are synthesized in the laboratory. No, they do not look like Hollywood actors with prostheses. And we do not know whether alien life forms use the alternative biomolecules that chemists cook up. For that, we must go to other worlds to look.
And no place is better than Mars.
[1] Piccirilli, J. A., Krauch, T., Moroney, S. E., Benner, S. A. (1990) Extending the genetic alphabet. Enzymatic incorporation of a new base pair into DNA and RNA. Nature 343, 33-37.
[2] Hoshika, S., Leal, N. A., Kim, M.-J., Kim, M.-S., Karalkar, N. B., Bates, A. M., Watkins Jr., N. E., SantaLucia, H. A., Meyer, A.J., DasGupta, S., Piccirilli, J. A., Ellington, A. D., SantaLucia Jr., J., Georgiadis, M. M., Benner, S. A. (2019) Hachimoji DNA and RNA. A genetic system with eight building blocks. Science 363, 884–887.
[3] Sefah, K., Yang, Z., Bradley, K. M., Hoshika, S., Jimenez, E., Zhu, G., Shanker, S., Yu, F., Tan, W., Benner, S. A. (2014) In vitro selection with artificial expanded genetic information systems (AEGIS). Proc. Natl. Acad. Sci. USA 111, 1449-1456.
[4] Zhang, L., Yang, Z., Sefah, K., Bradley, K. M., Hoshika, S., Kim, M.-J., Kim, H.-J., Zhu, G., Jimenez, E., Cansiz, S., Teng, I.-T., Champanhac, C., McLendon, C., Liu, C., Zhang, W., Gerloff, D. L., Huang, Z., Tan, W.-H., Benner, S. A. (2015) Evolution of functional six-nucleotide DNA. J. Am. Chem. Soc. 137, 6734-6737.
[5] Zhang, L., Wang, S., Yang, Z., Hoshika, S., Xie, S., Li, J., Chen, X., Wan, S., Li, L., Benner, S. A., Tan, W. (2020) An aptamer-nanotrain assembled from six-letter DNA delivers doxorubicin selectively to liver cancer cells. Angew. Chem. Int. Ed. 59, 663-888.
[6] Kimos, M. D., Khudyakov, I. Y., Alexandrushkina, N. I., Vanyushin, B. F. (1977) 2-Aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature 270, 369-370.
[7] Döring, V., Mootz, H. D., Nangle, L. A., Hendrickson, T. L., de Crécy-Lagard, V., Schimmel, P., & Marlière, P. (2001). Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science, 292(5516), 501-504.
[8] Marlière, P., Patrouix, J., Döring, V., Herdewijn, P., Tricot, S., Cruveiller, S., … & Mutzel, R. (2011). Chemical evolution of a bacterium’s genome. Angewandte Chemie, 123(31), 7247-7252.


Pingback: The Shadow Biosphere – The Primordial Scoop