Cite as: Benner, S. A. (2021) “The Shadow Biosphere”. Primordial Scoop, 2021, e0429. https://doi.org/10.52400/RPOH4426
A remarkable trio of papers today appeared in Science magazine1-3 (links: 1, 2, 3), which sees itself as the premier venue for scientific research on planet Earth. They report that some life on Earth does not have DNA with the standard four bases, adenine, thymine, guanine, and cytosine. Instead, some life (specifically, some viruses that infect various kinds of bacteria) has DNA that uses aminoadenine, thymine, guanine, and cytosine.
A picture is worth a thousand words. By adding an amino group to adenine, this novel DNA converts the Watson-Crick adenine:thymine base pair joined by two hydrogen bonds into the aminoadenine:thymine base pair, joined by three.
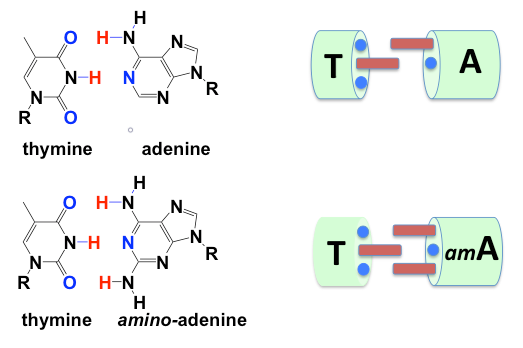
But this discovery not actually novel. As the Science papers point out, aminoadenine was first reported in a cyanobacteria virus genome by a team working in Russia in 1977;4 Ivan Khudyakov who participated in the 1977 discovery also coauthored one of the papers. That report was published in Nature, which also sees itself as the premier venue for scientific research on this planet (which means that they also do the embargo schtick).
But not actually novel. As all of the Science papers point out, aminoadenine was first reported in a cyanobacteria virus genome by a team working in Russia in 1977;4 Ivan Khudyakov, who was part of that 1977 study, also participated in this work. That report was published in Nature, which also sees itself as the premier venue for scientific research.
“So why are we talking about this now? What about this is new?” I was asked this by every reporter who called in for a quote.
For me, a professional who has artificially changed the DNA alphabet, lots of reasons. The teams with the French corresponding authors (Philippe Marlière, Pierre Alexandre Kaminiski), and the teams with American (Huimin Zhao) and Chinese corresponding authors (Yan Zhang and Suwen Zhao) did an excellent job of putting this new kind of DNA in context.
They figured out how aminoadenine was biosynthesized. That story is by itself worth a Single Scoop on this blog.
They identified mechanisms by which the virus might benefit from having aminoadenine in its DNA instead of adenine. Another Scoop.
They did a brilliant analysis of the evolution of the enzymes that make aminoadenine triphosphate and the polymerases that use aminoadenine triphosphate, showing co-evolution. This work showed that many viruses infecting many kinds of bacteria use aminoadenine, including those living in soil, fresh water, and sea water.
For me, this also has personal interest. One of these paper’s authors, Huimin, reminded me that he and his student, Yifeng Wei, got the idea to do this research after they saw a 2019 lecture of mine in Singapore on our artificial DNA. Afterwards, they questioned me about the possibility of similar things in nature. I directed them to the 1977 Russian paper in Nature. My life has now been worthwhile.
But even without personal involvement, we have good reason to see these reports as ground breaking. It goes under the title “The Shadow Biosphere”. About 15 years ago, Paul Davies, Charles Lineweaver [5], Carol Cleland, Shelley Copley [6], and others wondered whether Earth might have life living among and around us, unrecognized because of how we look for life. The historical example relates to the invention of the microscope, which changed our ability to look at the biosphere. Immediately, people using the microscope discovered a previously unsuspected world of micro-organisms.
Another example comes from work by Carl Woese and George Fox. They used technologies then emerging in the 1960’s to analyze the RNA molecules in living systems. They realized from these molecular analyses that an entire unrecognized kingdom of life exists, the Archaea. Paul Davies, Felisa Wolfe-Simon, and others sought such life in desert varnish, arsenic-laden springs, and other odd places on this planet.
Today, we look for life almost exclusively by molecular analyses. We take a sample from a hot spring, a swamp, or a large intestine, dump in primers to PCR amplify the DNA molecules that encode pieces of the ribosome (the machine that makes proteins). The PCR makes many copies of that DNA, and those copies are then sequenced.
All simple enough for polymerases that have evolved to handle adenine, thymine, guanine, and cytosine. But what if they do not amplify well DNA built from aminoadenine, thymine, guanine, and cytosine? We would miss that DNA, just as humankind for millennia missed the microbial biosphere because they did not have a microscope. What life we find is determined by the tools that we use to find it.
This is especially the case for hot springs. Normally, people think that when organisms evolve to live at high temperatures, they evolvd to have genomes rich in G: C pairs. Why? Because G:C pairs are already held together by three hydrogen bonds, and form a more stable duplex than A:T pairs.
But not so fast. Would not those organisms also have a reason to evolve to use aminoadenine:T pairs? After all, aminoadenine:T pairs are also joined by three hydrogen bonds. They are also more stable.
We have long wondered whether a shadow biosphere that uses some of our unnatural pairs might exist beneath our feet on Earth, unobserved because PCR that targets standard DNA would not amplify non-standard DNA well. And now the possibility has been made real.
And for Mars? Why not? We know how to look for life on Mars. We have reason to think that life emerged on Mars. Astrobiologists are inclined to think that life would “um … find a way” to persist after it emerged.
And yet NASA is not at this moment looking for the biosphere in the shadows of Mars in any way likely to find it. SpaceX will soon have the opportunity to look for life on Mars, but does not seem to be prepping itself with the latest technology to do so.
Perhaps it is time to get serious, to look for the shadow biosphere on Mars. Before it is too late.
- Pezo, V., Jaziri, F., Bourguignon, P.-Y., Louis, D., Jacobs-Sera, D., Rozenski, J., … Marliere, P. (2021). Noncanonical DNA polymerization by aminoadenine-based siphoviruses. Science, 372(6541), 520 LP – 524. https://doi.org/10.1126/science.abe6542
- Sleiman, D., Garcia, P. S., Lagune, M., Loc’h, J., Haouz, A., Taib, N., … Kaminski, P. A. (2021). A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes. Science, 372(6541), 516 LP – 520. https://doi.org/10.1126/science.abe6494
- Zhou, Y., Xu, X., Wei, Y., Cheng, Y., Guo, Y., Khudyakov, I., … Zhao, S. (2021). A widespread pathway for substitution of adenine by diaminopurine in phage genomes. Science, 372(6541), 512 LP – 516. https://doi.org/10.1126/science.abe4882
- Kirnos, M. D., Khudyakov, I. Y., Alexandrushkina, N. I., Vanyushin, N. I. (1977) 2-Aminoadenine is an adenine substituting for a based in S-2L cyanophage DNA. Nature, 270, 369-370.
- Davies, P. C., Lineweaver, C. H. (2005) Finding a second sample of life on Earth. Astrobiology, 5(2), 154-163.
- Cleland, C. E., Copley, S. D. (2005) The possibility of alternative microbial life on Earth. International J. Astrobiology, 4(3-4), 165-173.


Wonderful PrimordialScoop item! Excellent coverage of what indeed is a marvelous set of confirmations and elaborations for what has been in the shadow literature for nearly a half a century…
One of the important take home lessons is: even when it comes to the most basic chemical underpinning of present day’s life, no specific molecule is sacred. The reported discoveries support the idea that early in life’s origin (here and likely elsewhere) there were many “experiments” with diverse available chemistries and molecular mechanisms, later narrowing down evolutionarily to what we see today. So we should slow down on searches for “how did adenine specifically form prebiotically”. Perhaps a thousand molecular flowers bloomed at life’s inception, assemblies thereof just sharing the commandment “thou shalt reproduce”.
Yes Doron!
But the commandment was much more than “thou shalt reproduce”, rather something like “thou shalt be that which makes the meaning that is thyself out of the information that thou reproducest”!! Well, that would be a mouthful, even for the hypothetical Almighty, but that’s the point. One can excuse the mesmerising effect of DNA and information replication after the discovery of the base-paired structure, but the adamant and simplistic reduction of explanation to those terms (esp. Dawkins and his ilk) is a complete copout. There’s an elephant in the room.
I was pretty disappointed after Steve’s build up. I thought we were going to have an example of a clade of organisms that lived by using something other than the canonical “big 4 (ACGT)” base-alphabet. Not that I think we can’t learn from viruses. It is nice to see that there is a variant alphabet surviving in some host-parasite systems, but it is not an alternative that demands a different system of tRNAs, aaRS enzymes and variant ribosomes. As you point out, any mildly malleable chemical system will allow local variants, like ”I” instead of “1”, or “O” instead of “0”, but probably only surviving for 4 Gyr when the use of the variation is also involved in promulgating its own use.
There is no new idea in these results, just confirmation that there are minor variations in the systems of genetic information storage and transmission, just as there are variant codes.
This does not banish the elephant from the room. Surely the origin of interpreting information, rather than the particularities of its storage and replication, is where the key to our understanding of biology lies. Where did ANY system come from that was capable of interpreting the information it replicated? The means of interpreting information is provided by the products of the information’s interpretation. That means the phenotype is a self-organised dynamic structure, not some idle product of a dead mapping from one alphabet to another. It’s where the real biology lies. How did a code evolve from a state of absence of any interpretation of “genetic” sequences?
Sorry to disappoint. A virus is no less a life form just because it must eat ribosomes and glucose (much as you must eat glucose, while you provide your own ribosomes). However, given that constraint, you cannot reorganize translation. You are limited and how you can reorganize your genetic biopolymer In light of your virus-like dependency.
But what about bacteria? From my perspective, the interesting take away is the potential for a shadow biosphere. Polymerases evolved to handle G, T, C, and adenine will generally not handle as well G, T, C, and aminoadenine. Therefore, if we go looking for life by PCR using probes that target ribosomal genes, we will deplete from our amplicon pool ribosomal genes encoded by DNA containing aminoadenine. Therefore, we might expect a whole bacterial biosphere that uses aminoadenine hiding in the shadows because of how we look for it.
Thanks, Peter, for reminding us of some basics. Yes, there is way more to life than replication. Yes, as you point out the emergence of information processing is a central life puzzle piece. How was matter of any kind able to process information, to become cognitive? Replicating molecules are not cognitive.
At the risk of partially repeating myself, let me remind you all that there is a stability kind out there quite distinct to cognizantly-blind thermodynamic stability – dynamic kinetic stability (DKS). It’s a stability kind that depends on both the system AND its environment, so necessarily incorporates information on the environment. Accordingly, any DKS system, chemical or biological, is in some sense aware. So that’s the first step toward cognition – accessing the DKS state. Simplest DKS systems already manifest minimal cognition. In fact, thanks to the van Esch-Eelkema dissipative self-assembly work of recent years, a new dimension of chemical possibility has now opened up, a kinetic dimension. It helps explain why recently discovered chemical DKS systems exhibit biological-like characteristics, eg, self-healing, adaptive, communicative. I’ve offered a term for that new dimension – dynamic kinetic chemistry. Life is the extraordinary outcome of nature having explored dynamic kinetic chemistry for several billion years. We, in contrast, have been at it for less than a decade. It’s catch up time!